シン プレルナ助教/Singh, Prerna
Our research investigates how lipid flippases and ER architecture establish polarity and communication in plant cells. Using Physcomitrium patens as a model, we combine live-cell imaging, genetic manipulation, and biochemical reconstitution to uncover how lipid asymmetry and ER dynamics shape ROP nanodomain formation, cytoskeletal organization and developmental patterning. Ultimately, we aim to link molecular membrane remodeling to the emergence of cell polarity and multicellular coordination in plants.
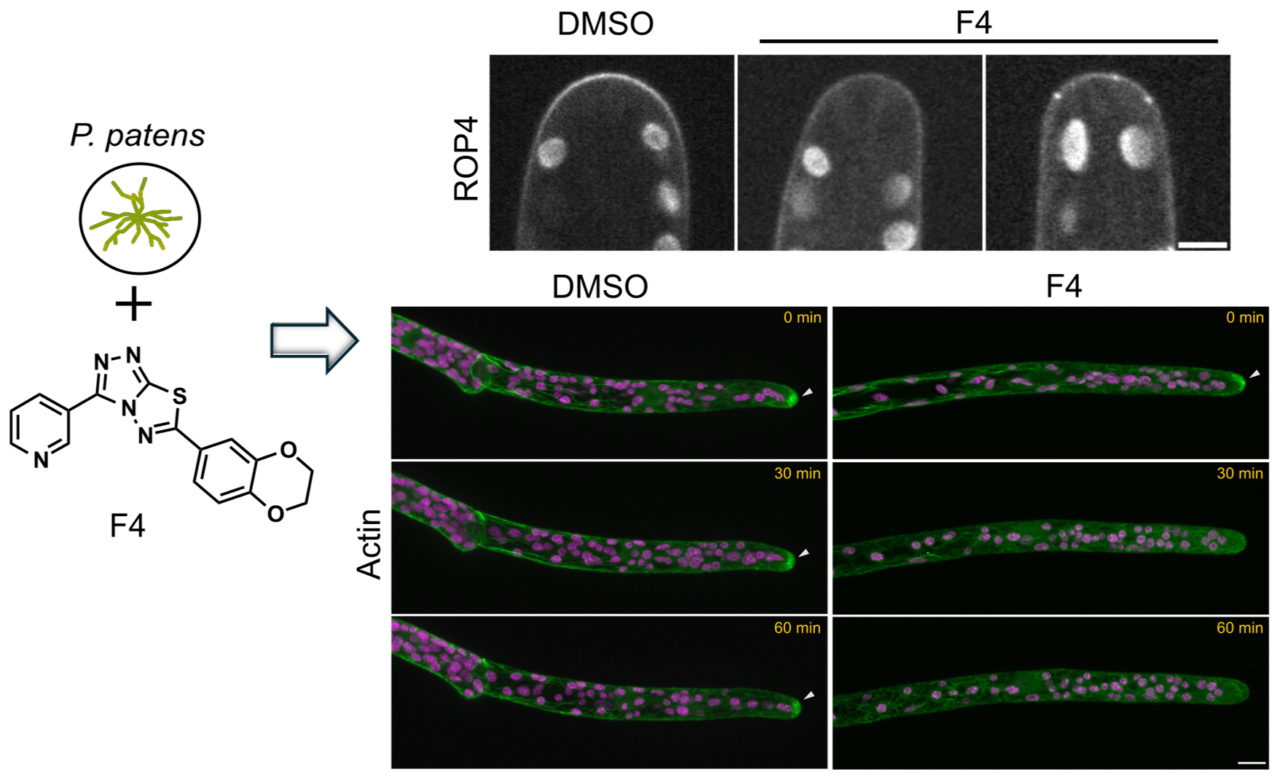
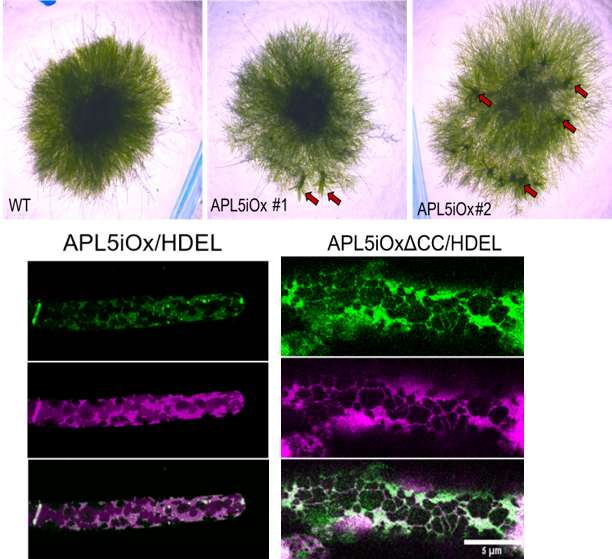
Cells are not static—they sculpt themselves from within. By exploring how lipids and membranes generate order, we uncover the physical logic behind life’s asymmetry. Join us if you’re curious about how molecules find direction, how cells build shape, and how simplicity gives rise to complexity.
参考文献
- Singh P, Jinno C, Zong H, Fujita T (2025) Asymmetry in the bryophyte Physcomitrium patens. Curr Opin Plant Biol doi:10.1016/j.pbi.2025.102760
- Singh P, Kadofusa N, Sato A, Naramoto S, Fujita T (2025) Novel small molecules disrupting polarized cell expansion and development in the moss Physcomitrium patens. Plant Biotechnol doi:10.5511/plantbiotechnology.25.0209a
細胞極性
閉じる
細胞骨格と膜の協調
閉じる
ケミカルバイオロジー
閉じる
脂質非対称性
閉じる
植物形態形成
閉じる
ヒメツリガネゴケ
閉じる

![[atmark]](https://www2.sci.hokudai.ac.jp/dept/bio/wp/wp-content/themes/sci-bio_2407/img/atmark.png)