TAKAHATA Shinya
Specially Appointed Lecturer
Gene Expression & Chromatin Dynamics
Department of Chemistry, Organic and Biological Chemistry

| Theme | Understanding the mechamnims of chromatin structure regulation and gene expression |
| Field | Molecular Biology, Epigenetics, Molecular genetics, Biochemistry |
| Keyword | Fission yeast, Budding yeast, Gene, Chromosome, Chromatin, Nucleosome, Transcription, Heterochromatin |
Introduction of Research
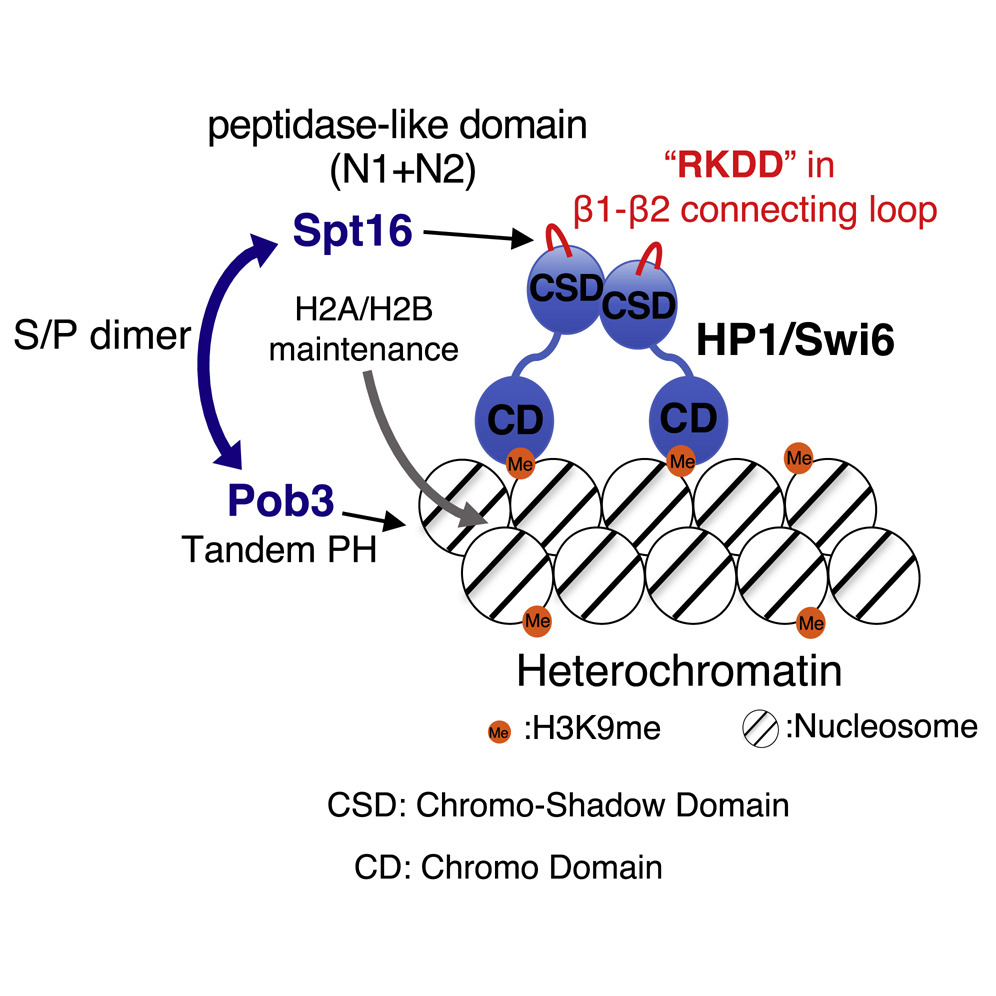
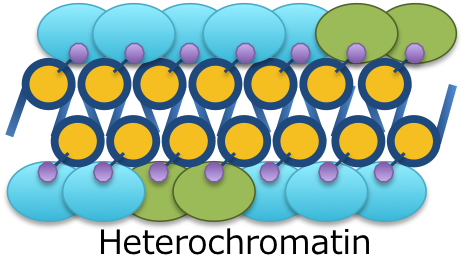
Genome contains numerous number of genes in eukaryote cells. Although there are 5000~ genes coded in yeast genome and 26000~ genes in human genome, all of genes are not equally expressed in cells. It depends on the situation in which the cells are placed. Gene expression is tightly regulated at several stages in living cells, and the landscape of gene expression pattern determines the cellular trait. Various factors are involved in this gene expression regulation, but the most important factor is a structural change of higher order conformation of chromatin. Gene expression is strongly repressed in a dense region of the chromatin structure on the genome, and gene expression is induced in a sparse region of the chromatin structure. Although it has been elucidated that it is a posttranslational chemical modification of histone protein to determine the sparseness and denseness of this chromatin structure, biological molecular reactions followed by the chemical modification at hitone tail is still obscure. The aim of our research is to determine biological supramolecules (proteins, RNA, and signaling chemicals) involved in chromatin structure regulation and to understand the mechanism for gene expression after transformation of chromatin structure at the molecular level.

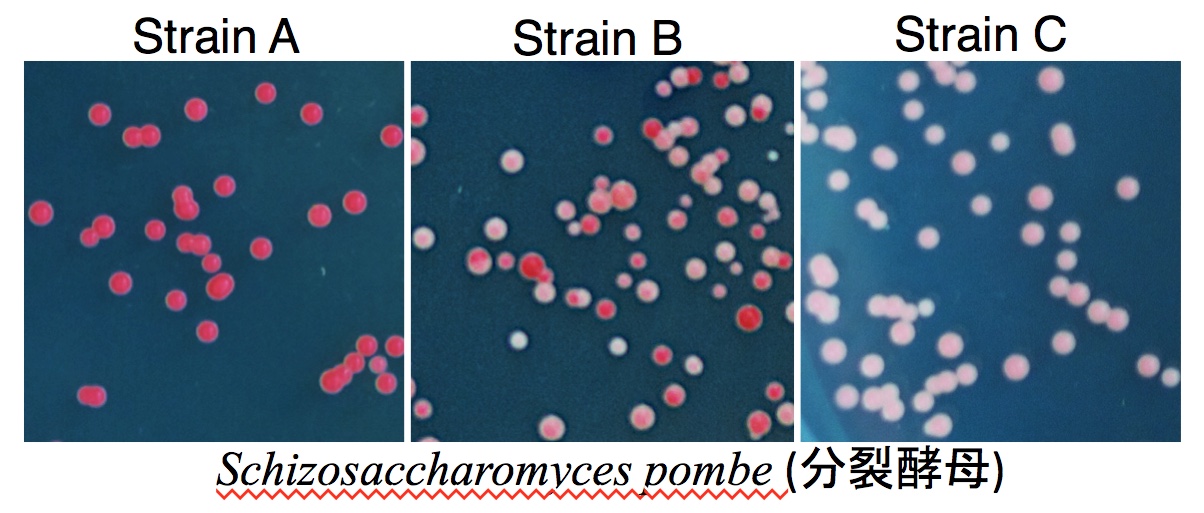
Representative Achievements
Related industries
| Academic degree | Ph.D. |
| Self Introduction | How genetic information is extracted from genome packaged in chromatin? It is a big mystery for our mankind. I am studying the molecular system that extracts genetic information from chromatin. |
| Academic background | 1993-1997 Faculty of Engineering, Kobe University B.E. 1997-1999 Biochemical Laboratory, SARAYA co. Ltd. Research Fellow 1999-2001 Division of Biological Science, Nara Institute of Science and Technology, M.S. 2001-2004 Graduate School of Integrated Science, Yokohama City University Ph.D. 2004-2005 Graduate School of Integrated Science, Yokohama City University PosDoc 2005-2009 School of Medicine, Univeristy of Utah, USA PosDoc 2009- current position |
| Affiliated academic society | The Molecular Biology Society of Japan, The Japan Society of Epigenetics |
| Room address | Faculty of Science, Building#2 2-203 |