WATAHIKI, MasaakiAssociate Professor
Auxin physiology and plant morphogenesis
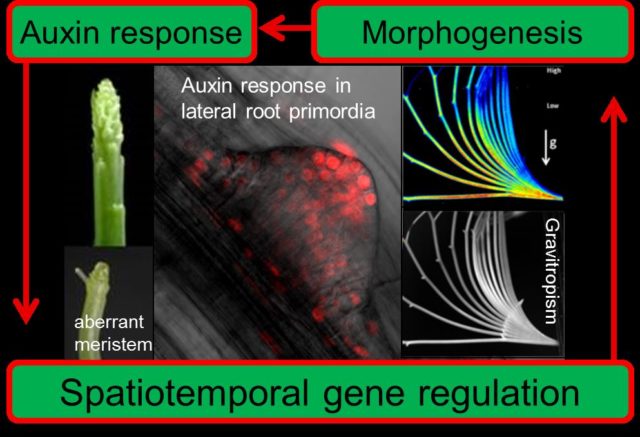
Plants are commonly considered as “Still life”, however, we just not able to sense their movement in our temporal sense. You may see the dynamic movement and development of plants in time-lapse movie. Plant hormone, “Auxin”, acts central role of these “Morphogenesis” in plant. This laboratory focused on “Early-Auxin Inducible Genes” and investigate the mechanisms how auxin works for making new organ (called “Organogenesis”). Arabidopsis is one of our experimental plants for forward genetics and reverse genetics but Potato and Carrot are participate in our project as well.
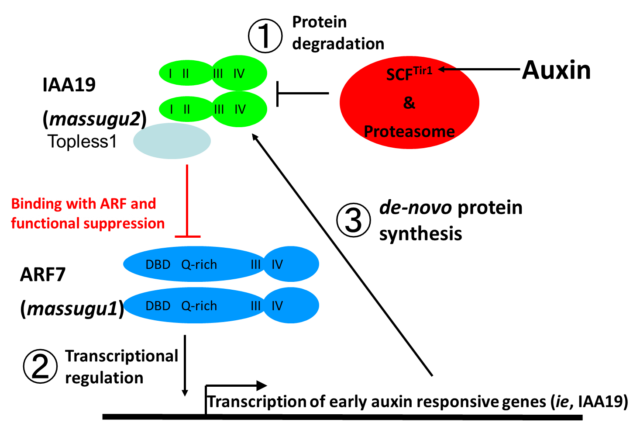
Auxin-insensitive mutants, massugu1 (msg1) and massugu2 (msg2) showed defect of gravitropism in etiolated hypocotyl. These mutant tend to grow straight (massugu in Japanese) irrespective to the direction of gravity. Both mutants are isolated by auxin-lanolin curvature test and reduces auxin sensitivity in hypocotyl (Watahiki and Yamamoto, 1997). msg1 is a recessive mutant and responsible gene encodes ARF7 transcriptional activator, while msg2 is a dominant mutant and encodes AUX/IAA19, which is induced by auxin and function as suppressor of transcription through topless protein. Dominant phenotype of msg2 caused by point mutation of domainII which is responsible for interaction with TIR1/AFBs. How dominant mutation suppresses auxin response? We hypothesize that auxin response of AUX/IAA19 is triggered by release of the suppression of ARF7 transcriptional activator (see “Binding with ARF and functional suppression” in figure below). ①Tir1/AFBs auxin receptor and AUX/IAA19 protein binds by small chemical, auxin (indole-3-acetic acid). AUX/IAA19 protein may ubiquitinated by SCF complex and ubiquitinated AUX/IAA degraded by 26S proteasome pathway. ② This decreases cellular (nuclear) concentration of AUX/IAA19 protein and leads to releases the suppression of ARF7 function. Activated ARF7 transcriptional activator target genes for expression. ③ However, ARF7 also activates AUX/IAA19 gene and de-novo protein of AUX/IAA19 suppresses ARF7. After all, auxin concentration controls cellular (nuclear) concentration of AUX/IAA19 negative and regulates transcriptional activity of ARF7。So, dominant mutation of domainII in msg2 mutant may explained that protein degradation of msg2 is not occurred by increase of cellular auxin level.
Although this model may apply if we do not consider of the time, the expression of AUX/IAA19 is transient (increase expression and then decrease) by application of auxin. Recent our study suggests that the auxin responsiveness of plant tissue can discriminate in basal state and stationary state. The organogenesis progresses by stepwise stages and auxin may contributes each steps. We speculates the conversion of “Basal state” and “Stationary state” could explain this stepwise progression of organogenesis. We named this conversion of auxin responsiveness as “Remodeling of auxin response” and investigates the molecular mechanisms.
Plants are auxotroph organisms and sustain our life as well as environment. Understandings of plant development will become increasingly important in future. My research aims for contribution of agriculture and sustainable environment from the academic knowledge and application of auxin research.
oin our Laboratory!!!
We are seeking motivated and well ambitious foreign students. Our group committed to teacing in Graduate school of Life Sciences, Biosystems Science Course. If you are looking for graduate student position, refer WEB site below and contacted with me. Hope your courage to investigate the fundamental mechanism of plant development with us.
Graduate School of Science, Biosystems Science Course HP
http://www.lfsci.hokudai.ac.jp/bs/en/
Higher Order Cellular Functions
http://www.lfsci.hokudai.ac.jp/bs/en/lab/#01
Information of Schlorships
http://www.oia.hokudai.ac.jp/prospective-students/scholarships/
About Hokkaido University
http://www.oia.hokudai.ac.jp/
http://grad.isc.hokudai.ac.jp/cgi-bin/index.pl?page=index&view_category_lang=2&view_category=0&allmenuopen=
References
- Kami C, Allenbach L, Zourelidou M, Ljung K, Schütz F, Isono E, Watahiki MK, Yamamoto KT, Schwechheimer C, Fankhauser C. (2014) Reduced phototropism in pks mutants may be due to altered auxin-regulated gene expression or reduced lateral auxin transport. Plant Journal. 77: 393-403.
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT (2007) Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiology, 144: 187-196.
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto K T (2004) MASSUGU2 Encodes Aux/IAA19, an Auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell, 16: 379-393.
- Harper R. M., Stowe-Evans E. L., Luesse D. R., Muto H., Tatematsu K., Watahiki M. K., Yamamoto K. T. and Liscum E. (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional growth in aerial tissues of Arabidopsis thaliana. Plant Cell, 12: 757-770.
- Watahiki M. K., Fujihira K., Tatematsu K., Yamamoto M. and Yamamoto K. T. (1999) The MSG1 and AXR1 genes of Arabidopsis are likely to act independently in growth-curvature response of hypocotyls. Planta, 207: 362-369.
- Watahiki M. K. and Yamamoto K. T. (1997) The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiology, 115: 419-426.
Website
WATAHIKI Laboratory We study for Auxin and Morphogenesis
Faculty
Faculty of Science
Department of Biological Sciences
Cell Structure and Function
Grad School
Graduate School of Life Science
Division of Life Science
Biosystems Science Course
Contact Information
Genome dynamics research center, West-GW
Email: watahiki ![[atmark]](https://www2.sci.hokudai.ac.jp/dept/bio/wp/wp-content/themes/sci-bio_2407/img/atmark.png) sci.hokudai.ac.jp
sci.hokudai.ac.jp