SATO, TakeoAssociate Professor
Plants need to sense and adapt to multiple environmental stress as they can’t move to other place. While we can’t see inside, they always change the cell activity though regulating protein function. In our laboratory, our major goal is to understand how plants regulate their growth in response to environmental stimulus. We are mainly use the Arabidopsis plants and studying the function of ubiquitin-proteasome system (UPS) for plant stress adaptation. UPS controls multiple protein stability through ATP-dependent and selective protein degradation by ubiquitin cascade and proteasome in eukaryote. Recently we isolated a novel ubiquitin ligase CNI1/ATL31, which regulates plant nutrient stress response called C/N (carbon/nitrogen balance) response, and revealed the ATL31 targets the 14-3-3 proteins for ubiqutiination and degradation in response to C/N status (Sato et al., 2009; Sato et al., 2011). In addition, we are studying the involvement of UPS to high CO2 adaptation and senescence since C/N is also important factor in these response. In addition to genetic and physiological analysis, we mainly use the proteomics methods such as immunoprecipitation and MS analysis to evaluate the protein interaction network and post-translational modification.
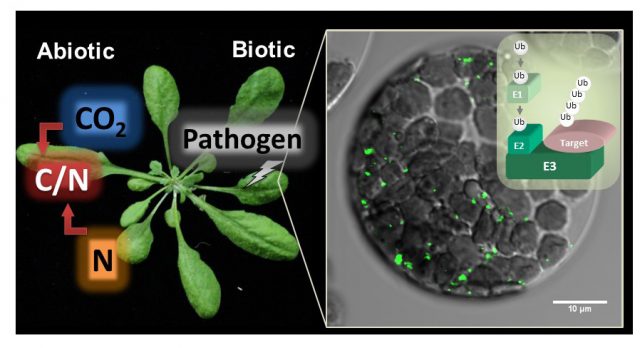
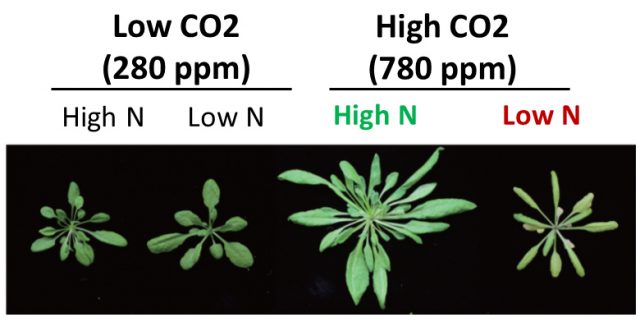
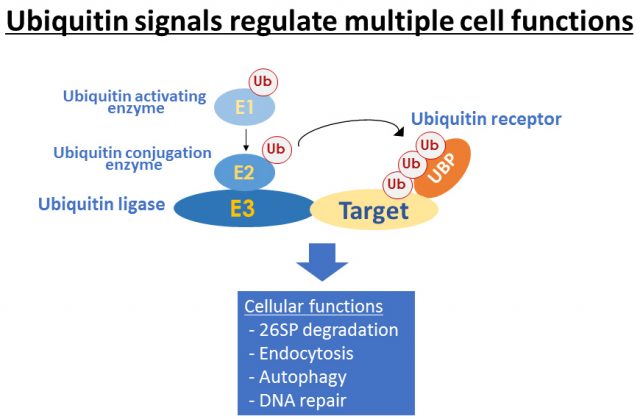
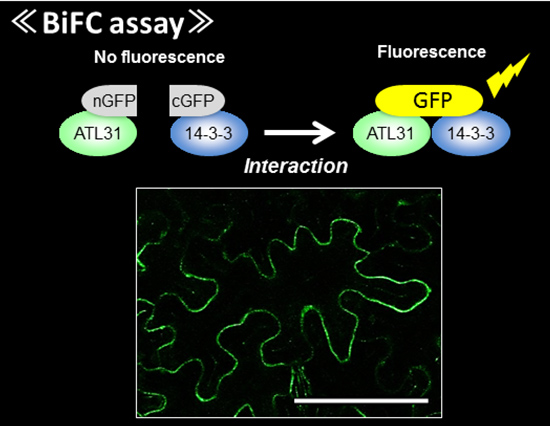
The information about plant nutrient adaptation mechanism including high CO2 response is getting more valuable for our life. We aim to reveal and provide the fundamental information for those subjects. Please feel free to contact me if you are interested in our project. I’m happy if I can meet and discuss with you.
References
- Luo Y, Takagi J, Claus LAN, Zhang C, Yasuda S, Hasegawa Y, Yamaguchi J, Shan L, Russinova E and Sato T*, Deubiquitinating enzymes UBP12 and UBP13 limit stabilize the brassinosteroid receptor BRI1. EMBO reports, 23: e53354. 2022
- Hasegawa Y, Huarancca Reyes T, Uemura T, Baral A, Fujimaki A, Luo Y, Morita Y, Saeki Y, Maekawa S, Yasudaa S, Mukuta K, Fukao Y, Tanaka K, Nakano A, Takagi J, Bhalerao RP, Yamaguchi J and Sato T*, The TGN/EE SNARE protein SYP61 and the ubiquitin ligase ATL31 cooperatively regulate plant responses to carbon/nitrogen conditions in Arabidopsis. Plant Cell, 34: 1354-1374. 2022
- Sanagi M, Aoyama S, Kubo A, Lu Y, Sato Y, Ito S, Abe M, Mitsuda N, Ohme-Takagi M, Kiba T, Nakagami H, Rolland F, Yamaguchi J, Imaizumi T* and Sato T*, Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A., 118: e2022942118. 2021
- Li X, Sanagi M, -5-, Schulze WX, Regina F, Stitt M, Lunn JE, Nakagami H*, Sato T* and Yamaguchi J, Protein phosphorylation dynamics under carbon/nitrogen-nutrient stress and identification of a cell death-related receptor-like kinase in Arabidopsis. Front. Plant Sci. 11: 377. 2020
- Sato T, Maekawa S, Konishi M, Yoshioka N, Sasaki Y, Maeda H, Ishida T, Kato Y, Yamaguchi J and Yanagisawa S, Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochem. Biophys. Res. Commun., 483: 380-386. 2017
- Yasuda S, Aoyama S, Hasegawa Y, Sato T* and Yamaguchi J, Arabidopsis CBL-Interacting Protein Kinases Regulate Carbon/Nitrogen-Nutrient Response by Phosphorylating Ubiquitin Ligase ATL31. Mol. Plant, 10: 605-618. 2017
- Lu Y, Yasuda S, Li X, Fukao Y, Tohge T, Fernie AR, Matsukura C, Ezura H, Sato T* and Yamaguchi J, Characterization of ubiquitin ligase SlATL31and proteomic analysis of 14-3-3 targets in tomato fruit tissue (Solanum lycopersicum L.). J. Proteomics, 143: 254-264. 2016
- Aoyama S, Huarancca Reyes T, Guglielminetti L, Lu Y, Morita Y, Sato T* and Yamaguchi J, Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol., 55: 293-305. 2014
- Yasuda S, Sato T, Maekawa S, Aoyama S, Fukao Y and Yamaguchi J, Phosphorylation of Arabidopsis Ubiquitin Ligase ATL31 Is Critical for Plant C/N-nutrient Response and Controls the Stability of 14-3-3 Proteins, J. Biol. Chem., 289: 15179-15193. 2014
- Maekawa S, Inada N, Yasuda S, Fukao Y, Fujiwara M, Sato T and Yamaguchi J, The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis, Plant Physiol., 164: 879-887. 2014
- Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB and Yamaguchi J, Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J., 68, 137-146. 2011
- Sato T, Maekawa S, Yasuda S, Sonoda S, Katoh E, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, Goto DB, Ikeda A and Yamaguchi J, CNI1/ATL31, a RING type ubiquitin ligase that functions in the Carbon/Nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J., 60, 852-864. 2009
Website
Faculty of Science and Graduate School of Life Science, Hokkaido University
Faculty
Faculty of Science
Department of Biological Sciences
Cell Structure and Function
Grad School
Graduate School of Life Science
Division of Life Science
Biosystems Science Course
Contact Information
Faculty of Science, Building #5 5-711
Email: t-satou ![[atmark]](https://www2.sci.hokudai.ac.jp/dept/bio/wp/wp-content/themes/sci-bio_2407/img/atmark.png) sci.hokudai.ac.jp
sci.hokudai.ac.jp
