Introduction of Research
Unveil molecular mechanism of primary active transporters by “seeing” their structures
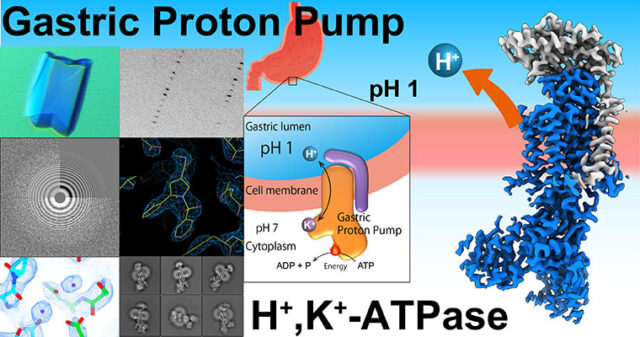
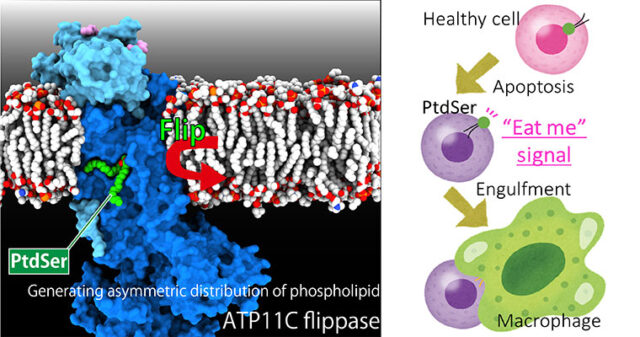
Asymmetric distribution of materials across the cell membrane is one of the important bases of the living system, which is generated by ATP-driven primary active transporters. Gastric proton pump generates pH 1 acidic environment in the stomach; Sodium pump is vital for the membrane potential especially important for neurons; phospholipid flippase translocate phospholipids acting as an #eat-me” signal for the apoptotic cells. The mechanism of how these target molecules work will be revealed by “seeing” their structures utilizing X-ray crystallography and cryo-EM single particle analysis.
Manipulate the transporters by chemical compounds
Design compounds that bind to target transporters, based on structural information of primary transporters that are vital for various physiological steps, and manipulate their activity – By collaborated with AI-driven de novo compound design and chemical synthesis, we will tackle drug development.
Create unknown primary transporters
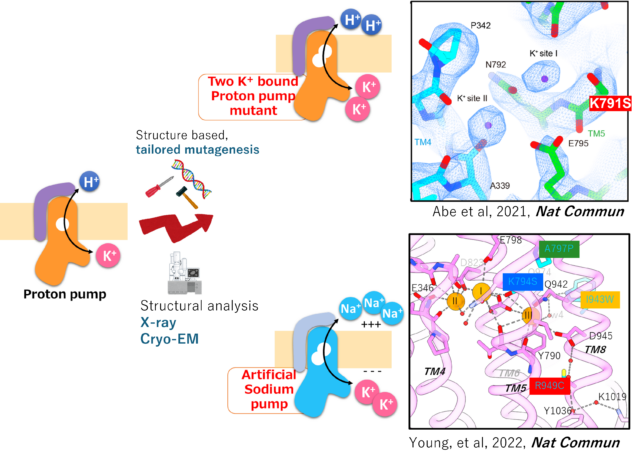
Modify existing active transporters based on the understood mechanism. By creating transporters that do not yet exist in this world, we will understand the necessary and sufficient for the transport mechanism.
Introduction of laboratory
This is a new laboratory established in 2024. Employing biochemistry, biophysics and structural biology, we will understand at the “chemical” level how active transporters work, to express their corresponding physiological phenomena by creating asymmetric distribution of various substrates.
