MURAKAMI Yota
Professor
Understand the regulatory mechanism of gene function
Department of Chemistry, Organic and Biological Chemistry

| Theme | Analysis of chromatin-mediated regulation of gene function |
| Field | Epigenetics, Molecular genetics, Genetics and dynamic regulation of genome, Biological chemistry |
| Keyword | Chromatin, Gene, Chromosome, Transposons, Genome, Centromere |
Introduction of Research
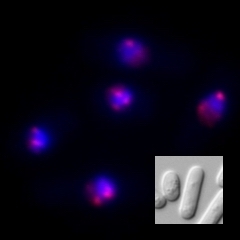
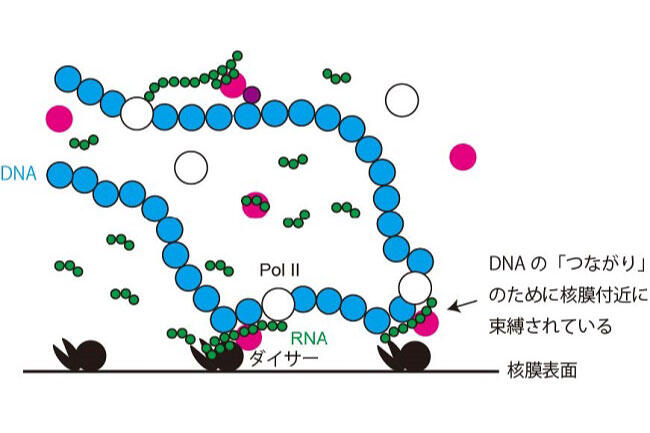
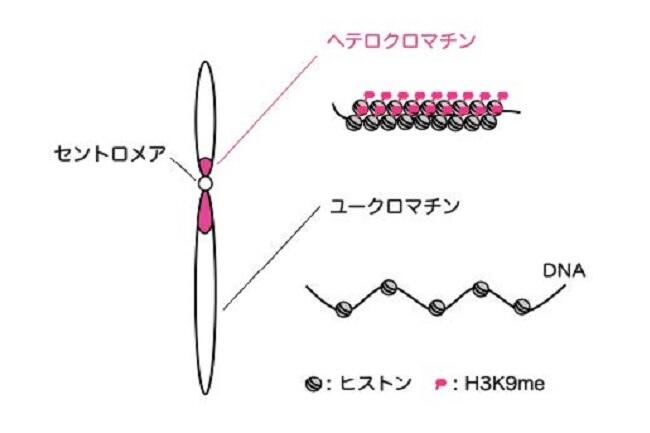
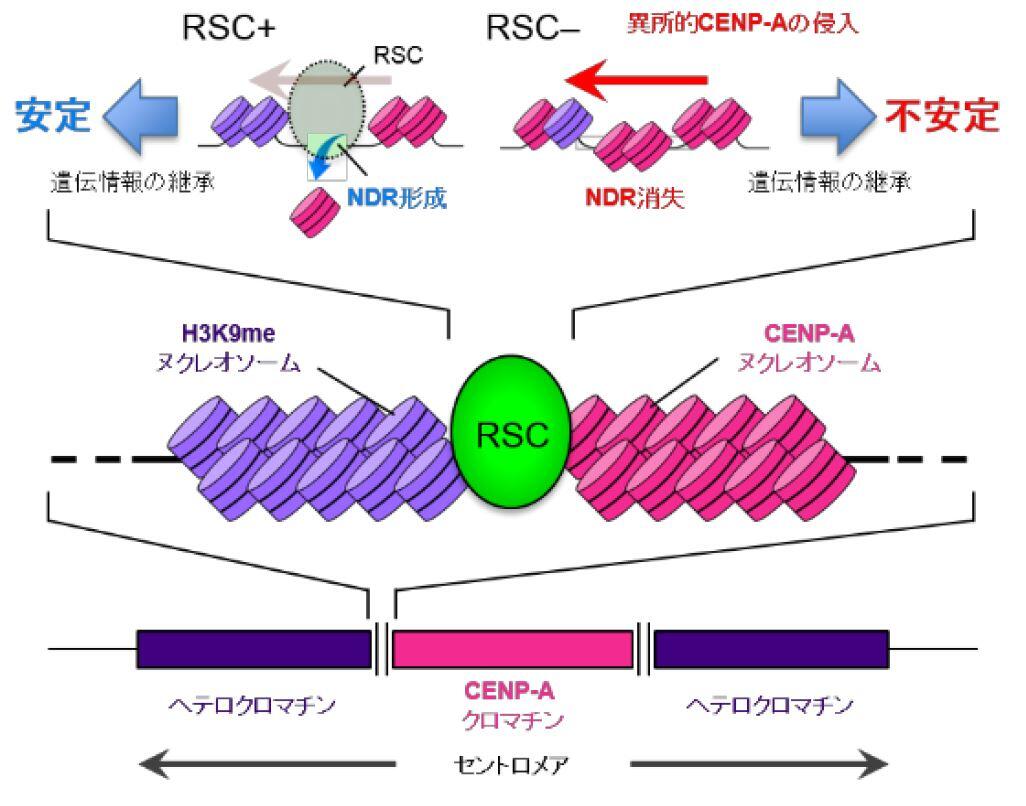
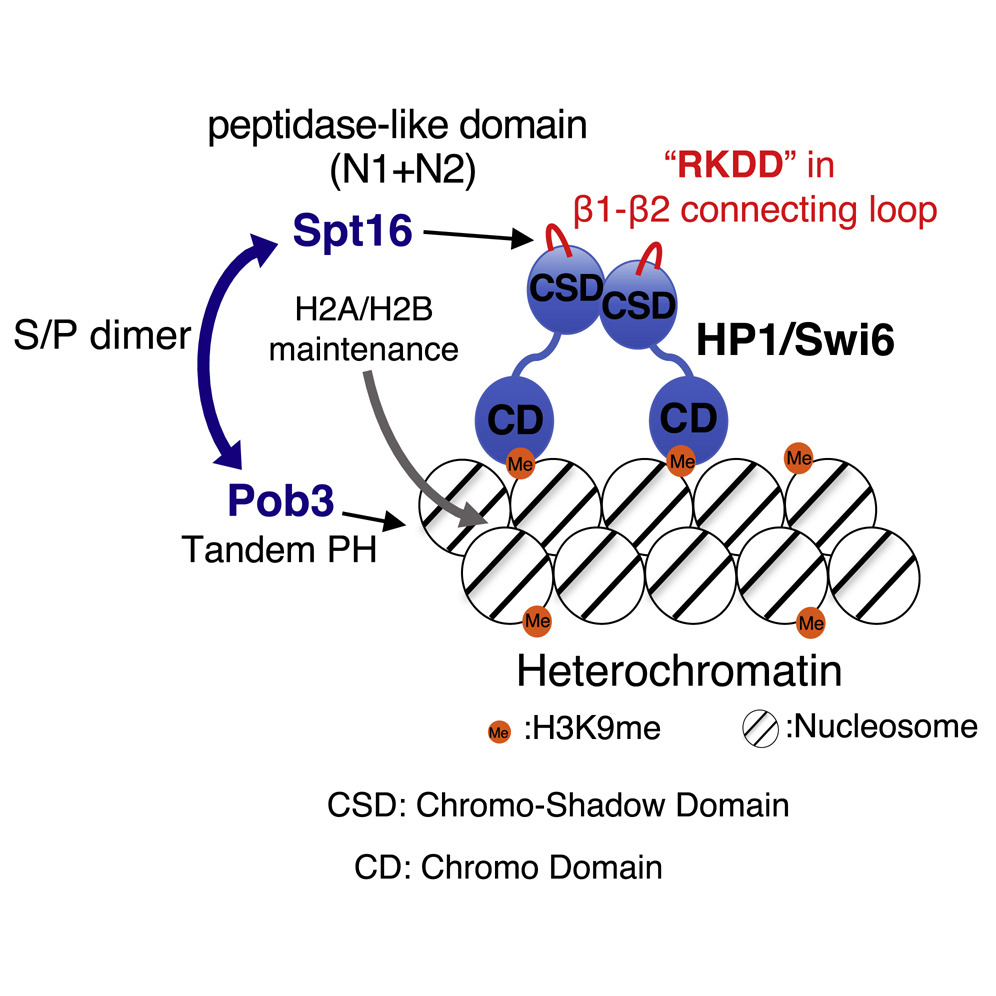
Long DNA, which stored an enormous amount of genetic information, is rapped around histone proteins, further folded into a specific structure called “chromatin” and packaged in the nucleus of the cell. Recent study revealed that the chromatin structure determined the specific gene expression pattern of various kinds of cells, which is stably maintained during cell divisions. This kind of regulatory system, in which gene expression is regulated without changing DNA sequences, is called “epigenetics”. Epigenetics is not only involved in cell fate determination, but also in oncogenesis and senescence. It is also important in the construction of iPS cells with applications to regenerative medicine. Importantly, the regulation of epigenetic gene expression is directly controlled by chromatin. We are interested in a special chromatin structure, heterochromatin. Heterochrimatin is not only associated with epigenetic regulation of gene expression, but is also deeply involved in the maintenance of genetic information, thus making it an important and complex chromatin structure.
We use the fission yeast as a model because its chromatin structure is similar to that of higher eukaryotes, although it is much simpler. In addition, mutants can be easily created in fission yeast. Various genes affecting the morphology and function of heterochromatin have been identified through the isolation and characterization of fission yeast mutant strains with abnormalities in its heterochromatin. We are aggressively analyzing several genes in relation to when, where, and how these function, including their reciprocal relationships. A variety of methods are being used to analyze its protein products, including the functional analysis, structural analysis of changes in heterochromatin structure using mutant strains, and the intracellular localization of each protein using fluorescence optics.
The concepts of epigenetics and heterochromatin are closely linked to cancer, senescence, and regenerative medicine; and have garnered attention from numerous researchers, thus making it a competitive field of research. However, it is a complex subject and research on it has just begun. With this background, we are proud of the results that we have achieved thus far. However, our goal is not just to win the race but to be one step closer to the mysteries of life, using our curiosity to endlessly conduct research.
Representative Achievements

| Self Introduction | I was born in Osaka, pure "Kansai-jin". Recently, I start running and dream to achieve "sub 3.5" in full marathon. |
| Academic background | 1981 Awarded the degree of BSc at Faculty of Science, Kyoto University. 1983 Awarded the degree of Msc at Faculty of Science, Kyoto University. 1987 Awarded PhD in Biological Physics at Faculty of Science, Kyoto University. 1987-1990 Post doctoral fellow at Inst. for Virus Research, Kyoto University 1990-1993 Post doctoral fellow at Sloan-Kettering Institute, NY, USA 1993-2009 Associate Professor at Inst. for virus Research, Kyoto University. 2009-present the present post |
| Room address | Faculty of Science, Bld #7 2007-02-08 |